How sensitive is the Guide-it SNP Screening Kit to the presence of mutations close to the site of the nucleotide substitution being assayed?
How sensitive is the Guide-it SNP Screening Kit to the presence of mutations close to the site of the nucleotide substitution being assayed?
The Guide-it SNP Screening Kit is highly sensitive to the presence of mutations in the genomic target sequence surrounding the site of the substitution being assayed. The displacement oligo and the flap-probe oligo anneal to the region immediately adjacent to the SNP, and the presence of unknown mutations in that region would affect the annealing of the oligos with the PCR product and the formation of the double-flap structure.
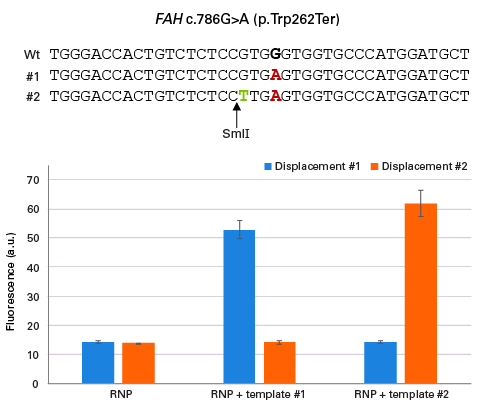
Sensitivity of the Guide-it SNP Screening Kit to mutations involving the genomic target sequence. The c.786G→A substitution in the FAH gene was engineered in hiPSCs by RNP electroporation together with either of two HR templates; one HR template (#1) encoded a sequence which included only the G→A substitution (in red), another template (#2) included an additional G→T substitution (in green) which generated a new SmlI restriction site (allowing detection of the substitution via RFLP). Two different displacement oligos were designed for both scenarios (i.e., G→A substitution vs. G→A and G→T substitutions in tandem), and used to assay samples obtained from the various edited cell populations. The oligo designed to assay the G→A substitution (displacement #1) only generated a strong signal for the sample electroporated with template #1. The complementary result was true for the displacement oligo designed to assay the two substitutions. These results show the sensitivity of the assay towards the sequence surrounding the edited base.
