Are there any limitations or special considerations regarding what target sequences can be analyzed with the Guide-it SNP Screening Kit?
Are there any limitations or special considerations regarding what target sequences can be analyzed with the Guide-it SNP Screening Kit?
The only design limitation one must consider when designing the probes is that the displacement oligo must not encode the sequences 5'-GGAGn-3' or 5'-GGAGNn-3' at its 3' terminus (where N can be any base, and n is the extra noncomplementary base that depends on the substitution being assayed). Displacement oligos containing these sequences will generate a background signal in the enzymatic assay. In these scenarios it is advisable to redesign the assay to target the opposite strand of the PCR product, such that the displacement oligo doesn't encode the restricted sequence at its 3' terminus.
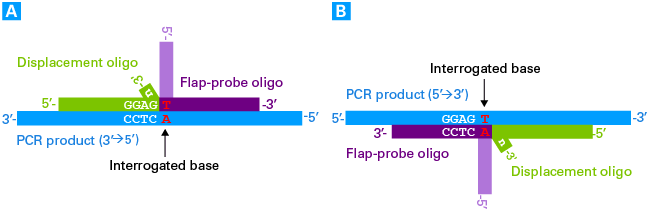
Example of a limitation in the design of the displacement oligo. In this scenario, the displacement oligo designed to hybridize with the PCR product (in the 3'→5' orientation) encodes the restricted sequence at its 3' terminus (Panel A). A viable alternative (Panel B) targets the opposite strand of the PCR product (in the 5'→3' orientation), avoiding the inclusion of the restricted sequence at the 3' terminus of the displacement oligo.
